1 Introduction
1.1 Historical Significance of the Cross-Bridge to Muscle Physiology and Kinesiology
A fundamental aspect of Kinesiology is centered upon understanding the mechanisms of human motion. Ultimately, every human movement is a direct result of a molecular motor (like myosin) transducing the chemical energy of adenosine triphosphate (ATP) into a mechanical force. Understanding how human skeletal muscle can accomplish the coupling between biochemistry and mechanics has transcended any single field of science over the past century and has required a multi-disciplinary approach to reach the current understanding we currently have on the topic.
In the early 1900s, pioneers in the field of muscle physiology provided the foundational work in understanding how muscles work. During this time the field of biochemistry was unclear of what the exact mechanisms were that provided a muscle cell with the necessary energy for muscle to contract, which was often contributed to oxygen (or at least some form of an oxidative pathway) (Bassett 2002) or to “lactic acid” (Herzog et al. 2015). In 1920, A.V. Hill observed the liberation of heat during isometric tension of an isolated muscle (Hill and Hartree 1920) showing that muscular force can be produced without oxygen. His findings ultimately led him to a Nobel Prize in 1922 (“The Nobel Prize in Physiology or Medicine 1922,” n.d.). This finding was detailed further when Wallace Fenn demonstrated that when allowed to shorten, a contracting muscle liberates more heat than when held isometrically (now well known as the Fenn effect) (Fenn 1924). Subsequently, Bailey (1937) characterized the abundance of myosin in skeletal muscle and Engelhardt and Ljubimowa (1939) demonstrated that myosin was indeed responsible for the muscle’s enzymatic activity and consequently the heat production that was observed by Hill and Fenn during their isolated muscle studies years prior. However, despite these early advances there was still no explanation for how a muscle (let alone a myosin molecule) could use ATP to produce force.
The next major breakthrough propelling the field closer to an answer for the mechanism of molecular force production came in 1954 when a pair of Nature articles from Huxley & Hanson and Huxley & Niedergerke independently provided evidence for muscle contraction via the “sliding filament theory” (H. Huxley and Hanson 1954; A. F. Huxley and Niedergerke 1954). Here, the two research groups describe the ability of myosin in the form of thick-filaments to associate with actin filaments causing sarcomere shortening (i.e. contraction) based on structural observations of contracting myofibrils and single muscle fibers. Even so, the sliding filament theory in of itself is not a mechanism of contraction. In 1957, Andrew Huxley provided the first mechanistic hypothesis about how the relative sliding of filaments could occur based upon modeling myosin as a biased Brownian ratchet. This proposal provided the initial groundwork for the modern-day “cross-bridge theory” of muscle contraction (HUXLEY 1957).
After the emergence of these structural perspectives of the actomyosin cross-bridge system biochemists began attempting to align their observations from solution kinetic studies to further explain the structure function relationship of the cross-bridge providing early kinetic schemes of the rates of the various mechanochemical steps/states of the cross-bridge model (Lymn and Taylor 1971). Since the completion of these foundational studies research into the field of muscle physiology has exploded as technology has advanced temporal and spatial limitations of instrumentation providing unprecedented details into muscle myosin structure and function which has provided a relatively deep understanding (versus the humble origins) of how the cross-bridges cycle from both a biochemical and structural perspective.
1.2 The Modern Cross-Bridge Cycle
The modern cross-bridge cycle is a mechanochemical system which describes the coordination between myosin’s enzymatic biochemistry and structural conformations. Different structural conformations are related to certain biochemical states and influence myosin’s affinity for the hydrolysis products and actin. As such, the cross-bridge cycle can be modeled as a summary of the current biochemical and structural conformations with a basic model being:
\[ ^{1)} AM_{post} ~ \implies ~ ^{2)} M_{pre} \cdot T~ \implies ~ ^{3)} M_{pre}\cdot D \cdot P ~ \implies ~ ^{4)} AM_{pre} \cdot D \cdot P ~ \implies ~ ^{5)} AM_{post} \cdot D \]
where A is actin, Mpost is myosin in a post-powerstroke conformation, Mpre is myosin in a pre-powerstroke conformation, T is ATP, D is ADP, and P is Pi.
1) Starting in rigor, myosin occupies a post-powerstroke position strongly bound to actin (Geeves and Holmes 1999; Sweeney and Houdusse 2010). 2) The binding of ATP to myosin causes a structural rearrangement in the active site which ultimately opens the upper and lower 50-kDa (Conibear et al. 2003; Kenneth C. Holmes et al. 2003) causing both the detachment from actin and resetting of the lever arm into a pre-powerstroke position (Nesmelov et al. 2011; Trivedi et al. 2015) . 3) ATP hydrolysis occurs constraining the myosin in the pre-powerstroke position. 4) When myosin attaches to actin there is a short lived strongly bound/load bearing state where myosin is pre-powerstroke with both hydrolysis products in the actin site (Llinas et al. 2015; Woody et al. 2019) . 5) The final step in the cycle is where myosin undergoes a powerstroke (i.e. lever arm rotation) and Pi-release which leaves the molecule in a strongly bound post-powerstroke state with solely ADP occupying the active site. Once ADP is released, myosin is left in rigor and the cycle restarts.
1.3 Powerstroke or Pi-Release? a biophysicists “chicken-or-egg” causality dilemma
While there are many details in the modern day cross-bridge cycle that are seemingly clear and well-established, there is a serious point of contention that remains in the literature to this day. The problem resides in the transition between the 3rd and 4th step in the model detailed in section 1.2. In one step, two key events are taking place - one mechanical and one biochemical. The mechanical powerstroke and biochemical release of Pi are collapsed into this single step without an explicit declaration of the timing or sequence relative to one other. Both these events are triggered by actin binding, but does the powerstroke precede Pi-release or does Pi-release “gate” the powerstroke?. This is the dilemma. Interestingly, the Powerstroke First and Pi-Release First models are both well supported in the literature. The divergence from a common mechanochemical scheme has divided the field for several decades and has ultimately limited the progress of fully understanding how myosin is able to transduce chemical energy into mechanical work as the powerstroke and Pi-release steps are key events in the transduction process. Ultimately, not understanding the precise order of the biochemical and mechanical events during the cross-bridge cycle prevents progress into the mechanisms underlying the molecular nature of energy conservation (1st law of thermodynamics) and limits our ability to develop pharmacological interventions to restore function to diseased, mutated, or compromised motors (HCM, DCM, fatigue, etc.). Moreover, in the emerging field of nanotechnology where researchers have been increasingly more interested in controlling biological motors understanding the precise ordering of steps in the transduction is paramount to the success of the field. For example, if you wanted to control myosin by preventing the powerstroke from occurring (and assuming a Pi-gating model of the cross-bridge) force production could be halted by trapping Pi in the active site. However, this approach would not work in a Powerstroke First model.
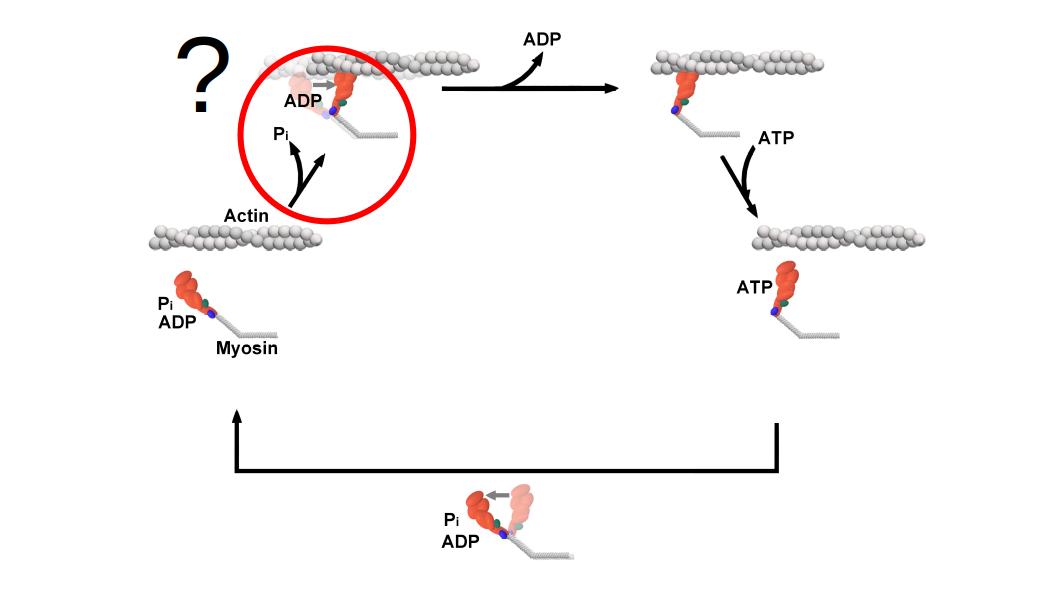
A typical model of the cross-bridge cycle. The powerstroke and release of Pi are often represented as occurring within the same step.
1.4 Q: How can the dilemma be resolved? A: By rigorously testing the current “unifying” theory set forth by structural biologists
In 2015, Llinas et al. (2015) performed x-ray crystallography on phosphate soaked myosin crystals and reported a new and never-before-seen myosin conformation that was posited to provide a unifying theory between the Powerstroke First and Pi-Release First models. This conformational state was aptly named the Pi-Release State (PiR State). Accompanying the PiR state, they provided a hypothesis regarding the structural sequence of Pi being released from myosin: 1) Myosin is in a Pre-powerstroke state (PPS) with Pi in active site, 2) the release of Pi from the active site into the so-called “phosphate release tunnel” causes slight re-arrangement into the PiR2 state (this is Powerstroke gating in their model), and 3) the final crystal structures detail Pi in the PiR1 state as the Pi is released into solution from the Pi-release tunnel. The subtly is that there are two different Pi-releases. A phosphate can be released from the active site into the Pi-release tunnel as well as be released from the Pi tunnel in myosin out into solution. Moreover, they state that the Pi-release from the active site into the Pi-release tunnel (PPS to PiR2 transition) is what gates the powerstroke and not the release from the tunnel into solution (see literature review for more details). Now, this is an elegant hypothesis that would explain away some of the most convincing functional data that supports the Powerstroke First model. Mainly, in coupled assays where the force of a muscle fibers is measured simultaneously with the rate of Pi-release (with a phosphate binding protein), or in the case with solution kinetic studies and FRET, the rate of the powerstroke is always measured to be far greater than any measured Pi-release rate. However, with the PiR state the structural biologists argue that a phosphate binding protein would be binding Pi in solution with the equivalent myosin structural state being PiR1 or later. Aka, the structural biologists are saying that an assay where a phosphate binding protein is used to measure the rate of Pi-release is actually measuring the wrong rate which makes the previous reports where the powerstroke was found to occur at faster rates an unfair comparison and that indeed the PiR2 to PiR1 is rapid and able to gate the powerstroke. However, this current “unifying” theory set forth in recent years remains to be rigorously tested and provides a testable hypothesis - If the transition of Pi from the active site into the PiR2 position gates the powerstroke, then can the powerstroke be prevented or slowed by occupying the active site with Pi?
1.5 Problem: the powerstroke is FAST. How do I see it? Solutions: Build a better mouse trap or get a slower mouse.
While the question is simple, “can the powerstroke be prevented or slowed by occupying the active site with Pi?”, devising a way to test this is not so simple for one main reason - the powerstroke occurs quite rapidly upon binding to actin (upwards of 5000/s) (Marco Capitanio et al. 2012; Woody et al. 2019). Being able to assess whether or not experimental conditions can affect the powerstroke is quite challenging. Ideally, these experiments will be conducted in a 3-bead laser trap assay which allows for the observation of single actomyosin binding events and through further analysis the displacement of every individual event can be measured and average displacements can be calculated and compared amongst the conditions. Furthermore, after the identification of the individual events, the trapping records can be ensemble averaged in order to measure the averaged rate of the transition from unbound to bound states. Experimentally, there are two ways that can be used to occupy myosin’s active with Pi. 1) By increasing the concentration of free Pi in solution the probability of Pi rebinding into the active site is increased, or 2) by using a mutation that prevents the release of Pi from the active site into the Pi-release tunnel (the S217A mutation in myosin V does just this) (Forgacs et al. 2009; Llinas et al. 2015). Really, the S217A mutation is just a “slower mouse” and affords the ability to use standard trapping techniques in order to test the “unifying” theory. The S217A mutation in myosin V has been shown to have an impaired rate of Pi-release of 30/s Llinas et al. (2015) which is far slower than the WT control myosin V (200/s). An event occurring at 30/s would on average take 33 milliseconds to be completed. If the release of Pi from the active does indeed gate the powerstroke there should be long delays between S217A myosin binding actin and the powerstroke (~33 milliseconds) which would readily be observable under a standard optical trapping setup that has millisecond time resolution. However, most WT myosins still have a relatively fast Pi-release rate when compared to the resolution of a standard optical trap which might still make the determination of the order of these events difficult. To overcome this, the Ultra-Fast Force Clamp (UFFC) has been used previously which provides sub-millisecond time resolution and allows for the observation of weak binding, an initial force bearing state, and the powerstroke itself in individual actomyosin binding events Woody et al. (2019). More simply, the UFFC is the “better mouse trap”.
1.6 Two steps forward, one step back
As presented there is seemingly a couple plausible experiments that can be performed to further assess the “unifying” theory of the powerstroke-first and Pi-release first model. However, there are currently not any free open source software (FOSS) projects whose goal is specifically for the automation of single molecule laser trapping data. This (perhaps) is one of the biggest obstacles that is limiting the field of single molecule biophysics. Without a community of software developers there are no standards for the analysis of our data and every new scientist (like myself) has to re-invent the wheel (or at least some version of it) from what they can piece together from the (sometimes very minimal) information provided by the methods section of an article. Unfortunately, I have no quantitative evidence supporting the limitations that a lack of analysis software is imposing on the field, but the evidence I do have is personal and anecdotal. This dissertation. As a graduate student I have identified and defended a research topic through my comprehensive exams and have even outlined in previous sections (see 1.4, 1.5) some of the scientific/experimental approaches that could serve as foundation to help progress the knowledge of the field on this given topic. However, even if I was handed all of the data needed to complete the project, I would not be able to answer the questions. Why? Because I do not have the right analysis programs/tools. Generally there are two common methods for analyzing single molecule laser trap data, 1) single molecule event identification (event picking) and 2) Mean Variance (MV) analysis (see Literature Review for more details on single molecule analysis techniques). Our lab primarily uses MV for analyzing single molecule data (Woodward et al. 2020; Unger and Debold 2019) and has had success with this method. While there are pros to using MV (see Literature Review) it may not be the best candidate to answer my specific questions. Mainly, MV does not identify individual events and as a result additional post-analyses cannot be run on the data, like ensemble averaging. With MV, a change in the magnitude of the displacement of the powerstroke could be estimated, but the rate of this transition could not be determined. This is a key question that needs to be answered for this project to be successful and so an event picking analysis program is needed. But, there are no available programs that implement this, that could be used easily, or that is FOSS. Sure, plenty of research groups have their own programs, but the source code is not available and details are limited (some authors do not even mention software/programming languages used) about how specific analyses work which makes replicating an analysis tricky and quite difficult and imposes a serious limitation to the reproducibility of work in the field.
1.7 Specific Aims
1.7.1 Aim 1: Develop software to automate the analysis of laser trap data
The current analysis that was in use by our lab (MV) was not sufficient to answer the questions presented here and answering these questions will ultimately help progress our understanding of how myosin converts chemical energy into mechanical work. An analysis program will be developed that can identify single molecule actomyosin interactions from raw trapping data with the ability to perform ensemble averaging in order to estimate the rate of the powerstroke (transition from unbound to bound). Moreover, the program will have a user-friendly graphical user interface (GUI) and be free and open source. The analysis will be written in the R programming language (which in of itself is FOSS) and the user interface built with Shiny (an R package that provides a web application framework). With both the analysis program and R language being FOSS, others may contribute/customize the analysis to their needs and improve the operating standards of single molecule trap analysis by offering community developed analysis solution. Additionally, this program will offer a practical, yet advanced, set of tools for other graduate students and research groups to easily implement in their own respective work. “Under the hood” the program will implement the current “best practices” of single molecule event detection by being able to access a robust network of R add-on packages written and developed by experts in their respective field (these R packages of course being well documented and well cited). Lastly, the program should not just be for analyzing single laser trap data records, but should be in fact a complete data analysis and data management tool that will offer easy-to-use tools to perform calibrations, data cleaning and management, analysis, as well as being able to perform project summary statistics. In essence, this program will be a tool that will completely automate the analysis of laser trap data from raw data to final figures.
1.7.2 Aim 2: Test the “slower mouse”. Determine if the S217A mutation in myosin V has a reduced displacement or rate of its powerstroke.
S217A is an ideal candidate to test the sequence of events surrounding the timing of the powerstroke and Pi-release in the 3-bead laser trap assay. The main advantage being that S217A effectively traps the γ-phosphate in the active site which has been shown to have a drastic effect on slowing the Pi-release rate as previously measured in solution kinetics studies (~30/sec). Our hypothesis is that if Pi-release gates the powerstroke, we should observe a prolonged (~33 milliseconds) delay (time period characterized by a decrease in variance of the trapping signal with no displacement) between the initial binding of myosin to actin and occurrence of the powerstroke. The resulting effects of the S217A mutation would be a reduced displacement (leftward shift in the displacement distribution compared to WT) and a slowed transition rate in the ensemble averaged data. However, if the powerstroke precedes Pi-release then the S217A displacement distributions, and ensemble averages should look identical to the WT. Additionally, performing this same experiment again with high (30mM) Pi in solution will provide an alternative methods for attempting to occupy the active site with Pi. As such, our hypothesis is that if Pi gates the powerstroke there should be a prolonged delay between the initial binding and powerstroke (but much longer in time for S217A vs WT due to slowed Pi-release rate). If the powerstroke precedes P-release then all 4 conditions WT 0mM-Pi, WT 30mM-Pi, S217A 0mM-Pi, and S217A 30-mM-Pi will have identical displacement distributions and ensemble averages.
1.7.3 Aim 3: Test the “better mouse trap”. Determine if fast chicken skeletal muscle myosin II has an altered displacement or rate of its powerstroke under high levels of Pi by analyzing data from the UFFC.
While the S217A provides an ideal test since the mutation has drastic affects on slowing the Pi-release rate, there are still limitations imposed by the setup of the standard optical trapping technique that prevent direct observation of myosin binding actin and the subsequent powerstroke. UFFC provides an increased time resolution and allows for the direct observation of the rate of a single powerstroke by a single molecule of myosin interacting with a single actin filament. This is in contrast to inferring an averaged “powerstroke” rate from the ensemble averaged data collected under a standard trapping protocol. Furthermore, the UFFC has the necessary time resolution in order to test a faster myosin isoform (chicken fast skeletal muscle myosin II) under no (0mM) and high (30mM) Pi which will provide additional data on a different myosin isoform helping to provide a more robust and inclusive answer to the proposed question. Our hypothesis is if Pi-release gates the powerstroke there should be a prolonged initial weak binding state between the binding of myosin to actin and the powerstroke under 30mM Pi compared to 0mM Pi, but the rate of the actual powerstroke should be unchanged.