6 Ultra-cool Conclusion
With the combined efforts of the work in Chapter 4 and Chapter 5 which together provide a seemingly robust, precise, and accurate analysis app which was used to analyze single molecule laser trap data under conditions high levels (30mM) of Pi and with our “slow mouse” S217A mutation our primary conclusions reached thus far supports the notion that the powerstroke proceeds Pi-release as we were unable to reject the null hypotheses - we saw no effects of the S217A mutation or high levels of Pi (or any interactions) on the absolute displacements or on the rate of unbound-to-bound transitions as revealed via the ensemble averages.
6.1 The “cherry-on-top”
The potential drawbacks of the data from Chapter 5 is the reliance on data being collected from a mutated myosin and the caveat that the unbound-to-bound transition is not a measurement of the rate of myosin’s actual powerstroke, but is instead the observable rate the reflects the underlying process of myosin’s powerstroke displacing glass beads through solution, a rate heavily damped by viscous drag forces. However, there exists the “better mouse trap” - the ultra-fast force clamp. Advisor and commitee members have graciously allowed me to include some of their data collected with the UFFC in this dissertation as the dataset is relevant to the main question attempting to be addressed (Powerstroke vs Pi-release first) here. The strength of the data provided by the UFFC offers a strong counter-argument to anyone concerned with any of the potential drawbacks to the concerns mentioned above in regards to Chapter 5 (not a WT myosin that exists in-vivo, indirect measure of transition rate), as UFFC provides a direct quantification/visualization of myosin’s powerstroke and affords the time-resolution to use a wild-type myosin that really exists in live cells and organisms.
6.2 Direct observation of myosin’s powerstroke
Looking at some of the raw data from the UFFC, several nanometer displacements (~4nm) were observed after the intial binding of myosin to actin. A small portion of raw data is shown in Figure 6.1. The baseline is defined by the wide triangular wave imposed by the QPDs stearing the beads rapidly back and forth to drive the beads at a fixed velocity and force. As the system is designed to keep a constant force on the beads, when an actomyosin interaction occurs causing the feedback to disengage, stopping the oscillation of the beads as the stiffness of the myosin will be exerting the forces required to clamp the force (shown in blue). After a brief dwell period, which is assumed to be a weak binding state, myosin progresses through its powerstroke (green) which is directly observed in the UFFC. As such, this is the only instrument fast enough to apply feedback to the actual mehanics of force generation at the level of a single molecule.
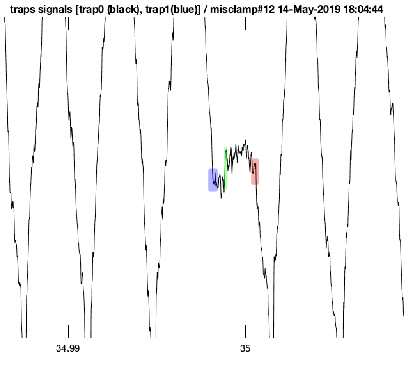
Raw data trace from the UFFC showing the intitial binding of myosin to actin (blue), direct observation of the powerstroke (green), and ATP induced dissociation (red). Data was collected at 1mM-ATP and 30mM-Pi. Time scale is presumably in seconds so the entirety of the event is ~2ms.
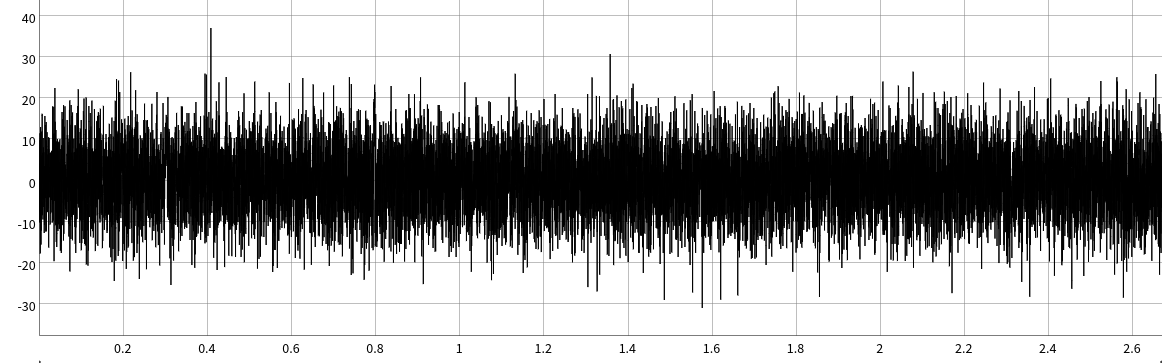
The ability for ultrafast to detect these single molecule events with a skeletal muscle myosin II, a very fast low duty ratio motor, at these of high ATP concentrations (1mM) is really just plain awesome. For context, this is a 2-second simulated data trace of what very short 2-10ms data would look like in the standard laser trap setup used in Chapter 5.There are 5 single molecule displacements in this 2 second simulation that are between 1-10ms in duration. Can you spot them all?
The raw data shown in Figure 6.1 displays a single binding event under high levels of phosphate. This is a similiar response and time-scale to the sequence of binding, dwell, and displacement as observed under no phosphate conditions which indicates that the initial steps in the process of force generation is un-affected by phosphate concentration which supports the powerstroke first model. The expectation in a Pi-release first would be that the addition of high levels of Pi, combined with the resitive load, would promote Pi to rebind to myosin’s active site after the initial release which would then prolong the initial dwell time prior to the powerstroke. A conceptual model of what a single molecule interaction might look like in the UFFC is shown in Figure 6.3 under a Pi-release first model.
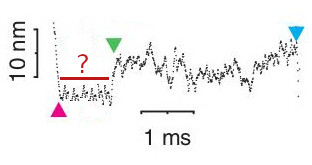
A possible conceptual representation of a single UFFC event under a Pi-release first model in high Pi concentration conditions. This effect was not observed.
6.3 Ensemble averages
With a similiar methodology to the ensembles average technique as described in previous chapters, UFFC events can be averaged and ensembled together. However, an important distinction between the ensemble averages between ultrafast and those shown in previous chapter with a standard laser trap is that the ensembles average shown here more accurately represents the average rate of myosin’s powerstroke since the stroke is being directly observed in the single events. For this reason the terminology is carefully chosen when describing the intial rates of the two different types of ensembles averages. The wording “unbound-to-bound transition” is used in Chapter 5 to indicate that we are privy to the notion that this rate is much slower than the true rate of the powerstroke. Figure 6.4 shows an ensemble averaged data trace from the ultrafast trap collected with fast chicken skeletal (full-length) myosin under high levels of 30mM-Pi. Similarly to the individual events, the ensembles show a rapid displacement after the initial interaction with actin that is un-affected by the addition of Pi into final solution conditions. As is evident in Figure 6.4, even on average in the ensembles the powerstroke occurs within several microseconds after the the initial actomyosin interaction. The timing of this observation is far greater than any known measured rate of Pi by several orders of magnitude providing strong evidence of the powerstroke processing Pi-release.
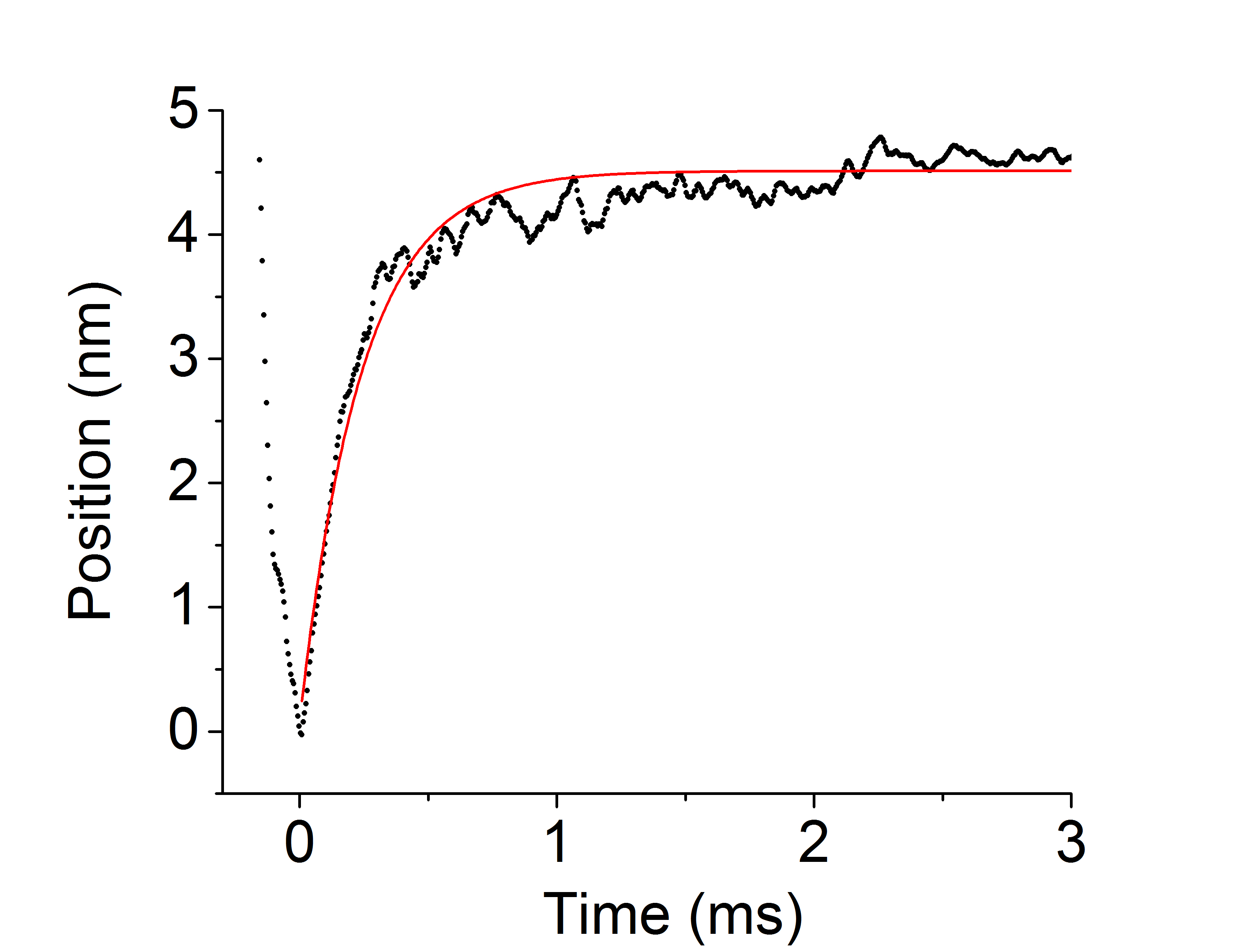
Ensemble average of UFFC data at 30mM-Pi. Myosin’s powestroke occured rapidly withing several hundred micro-seconds even under high Pi conditions and substantial resistive load.
6.4 Final Remarks
In Chapter 5 we used two independent approaches to test the relative timing of myosin’s powerstroke and Pi-release by using high levels of Pi in solution and using a mutation in attempts to promote Pi occupying the active site. In both cases myosin generated a powerstroke rapidly upon binding to actin as evidenced through the ensemble averages and did not alter the magnitude of the displacement. These data provide strong evidence that myosin generates its powerstroke when Pi is still in its active site which supports the notion of a powerstroke first model of myosin’s cross-bridge cycle. These results are in agreeance with recent data from the ultrafast optical trap (Woody et al. 2019) performed with cardiac myosin and with the ultrafast data that was showcased in this work with skeletal myosin and higher Pi concentrations. The magnitude of providing an answer to this question cannot be overstated as this is not just at the root of understanding how myosin transduces energy, but due to the conserved amino acid sequence in the active site of the myosin family tree and related molecule motors (Vale 1996; J. E. Walker et al. 1982) the answer to this question will provide insight at a fundamental level in a more general sense about how enzymes convert chemical energy into mechanical work in order to accomplish a wide variety of cellular tasks such as muscle contraction (Kenneth C. Holmes 1997), cell division (Zang et al. 1997), and intracellular cargo transport (Cooper 2000; Titus 2018). Most specifically to the field of muscle physiology and kinesiology this research is important and should be conducted because it falls in the direct lineage and can provide an answer to the question that originally motivated Hill, Fenn, Huxley, and Huxley - how do muscles work?